According to experts, thousands of lives could be saved after the discovery of a new type of prostate cancer.
Artificial intelligence has helped scientists discover a new form of the disease that could revolutionize the way it is diagnosed and treated in the future.
Their study has found that prostate cancer, which affects one in eight men during their lifetime, includes two different subtypes.
And the revelation could lead to personalized treatments for each individual patient depending on which type they have.
The team, led by researchers from the University of Oxford and the University of Manchester, used artificial intelligence (AI) to study changes in DNA in prostate cancer samples from 159 patients.


Stephen Fry, 66, was diagnosed with prostate cancer in 2018 but has since recovered. Last week he supported the #CatchUpWithCancer campaign and criticized the “deadly” delays tens of thousands of people have faced.
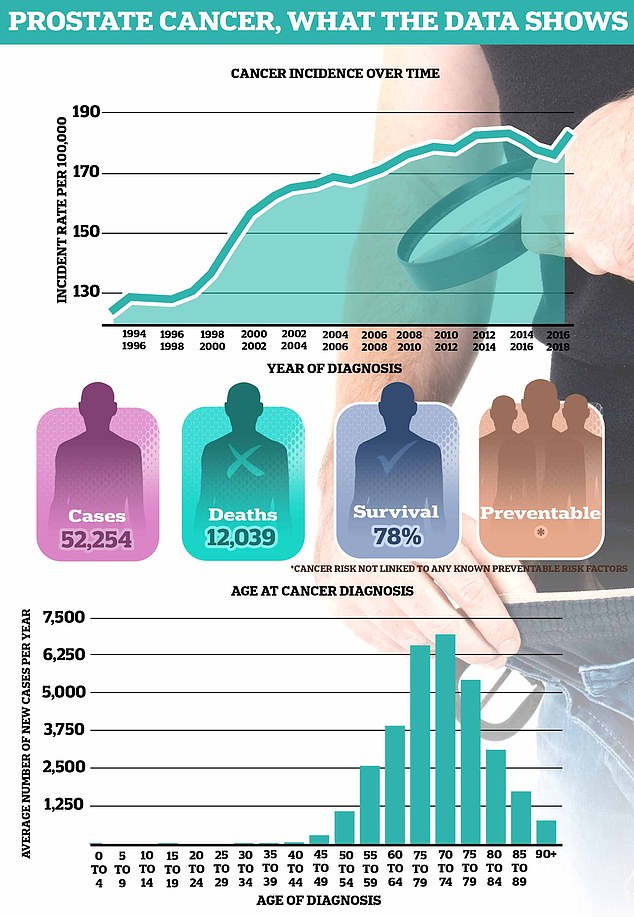

More than 52,000 men are diagnosed with prostate cancer each year on average in the UK, making it the most common cancer in men. Around 12,000 men die each year from this disease, the equivalent of one every 45 minutes.
They identified two distinct groups of cancer among these patients and were able to generate an “evolutionary tree” showing how each of them developed, ultimately converging on two distinct types of diseases called “evotypes.”
This is significant since until now it was thought that prostate cancer was just one type of disease.
Lead researcher Dr Dan Woodcock, from the University of Oxford, said: “Our research shows that prostate tumors evolve along multiple pathways, leading to two distinct types of diseases.”
“This understanding is critical as it allows us to classify tumors based on how the cancer evolves and not solely on individual genetic mutations or expression patterns.”
The researchers worked together as part of an international consortium called The Pan Prostate Cancer Group, created by scientists from the Institute of Cancer Research (ICR) and the University of East Anglia to analyze genetic data from thousands of prostate cancer samples in nine countries. .
The team’s collaboration with Cancer Research UK (CRUK), which funded the study, aims to develop a genetic test that, when combined with conventional staging and classification, can provide a more accurate prognosis for each patient, enabling treatment decisions. personalized.
Dr Rupal Mistry, Senior Director of Scientific Engagement at CRUK, said: “The work published today by this global consortium of researchers has the potential to make a real difference to people affected by prostate cancer.
«The more we understand about cancer, the more possibilities we will have of developing treatments to combat it.
“We are proud to have helped fund this cutting-edge work, which has laid the foundation for personalized treatments for people with prostate cancer, allowing more people to overcome their disease.”
Research Professor Colin Cooper, from the University of East Anglia’s Norwich Medical School, highlighted that while prostate cancer is responsible for a large proportion of all cancer deaths in men, a disease caused by prostate cancer is more common. that men die for, that for which they die.
This means that unnecessary treatments can often be avoided, sparing men side effects such as incontinence and impotence.
He added: “This study is really important because until now we thought that prostate cancer was just one type of disease.”
“But only now, with advances in artificial intelligence, have we been able to show that there are actually two different subtypes at play.
“We hope that the findings will not only save lives through better diagnosis and personalized treatments in the future, but can also help researchers working in other cancer fields better understand other types of cancer.”
Dr Naomi Elster, research director at Prostate Cancer Research, said: “These results could be the start of us taking the same ‘divide and conquer’ approach to prostate cancer that has worked for other diseases, such as breast cancer.’
The findings were published in the journal Cell Genomics.

