From old iPhones to discarded vapes, a shocking new report warns that our old technology is creating unmanageable amounts of e-waste.
The UN report reveals that in 2022, the world created 62 million tonnes of electronic waste, the equivalent of the weight of 6,000 Eiffel Towers.
Worryingly, global waste is increasing by 2.6 million tonnes each year and could reach 82 million tonnes by 2030.
What’s more, less than a quarter of this waste is recycled, despite containing billions of dollars’ worth of gold and rare minerals.
Kees Baldé, lead author of the report, said: “No more than 1% of the demand for essential rare earth elements is met by e-waste recycling. Simply put: the status quo cannot continue.

A shocking report reveals that the world generated 62 million tonnes of e-waste in 2022, including thousands of tonnes of discarded solar panels. This image shows piles of waste awaiting recycling at a center in Germany
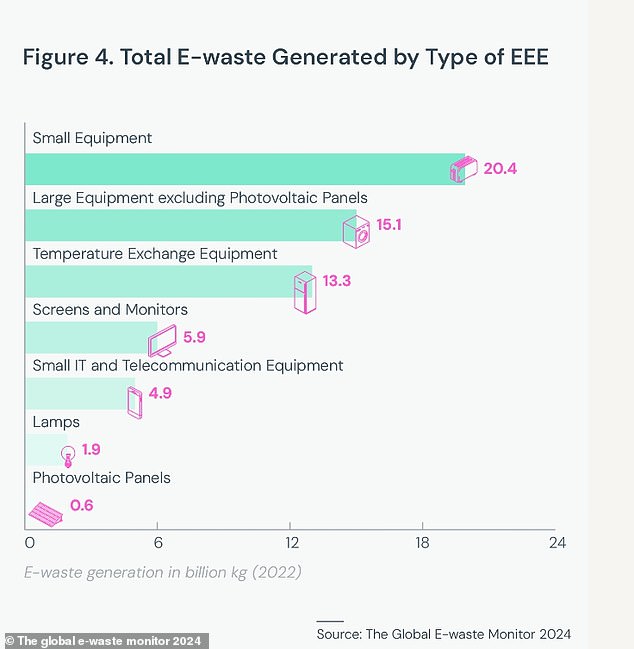

A third of the 62 million tonnes of e-waste generated in 2022 was classified as “small devices”, which includes everything from toys to microwaves.
A third of the 62 million tonnes of e-waste generated in 2022 was classified as “small devices”, which includes everything from toys to microwaves.
Another 4.6 million tonnes come from what the report calls “small IT and telecommunications equipment”.
This includes some of the most commonly used devices such as laptops, mobile phones, GPS devices and routers.
In the future, researchers suggest that solar panels could become one of the largest contributors to e-waste.
In 2022, only 600,000 tonnes of solar panels ended up as e-waste, but by 2030 this figure could reach 2.4 million tonnes.
Nikhil Seth, Executive Director of UNITAR (United Nations Institute for Training and Research), said: “Amid the hopeful adoption of solar panels and electronic equipment to combat the climate crisis and driving digital progress, the emergence of e-waste requires urgent attention. ‘
European countries are the largest producers of e-waste per capita, creating an average of 17.6 kg per person per year.
This is followed by Oceania with 16.3 kg per person, the Americas with 14.1 kg and Asia with 6.4 kg per person.
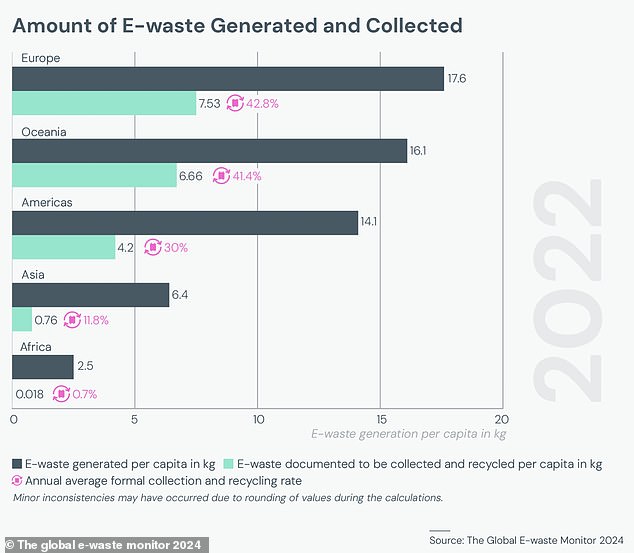

Europe and Oceania produce the most e-waste per person. However, they also have the most developed recycling infrastructure, meaning less waste ends up in landfills.


Cell phones and other telecommunications devices accounted for 4.6 million tons of electronic waste worldwide. This image shows phones collected for informal recycling in Ghana.
However, it is not only the amount of waste generated that is important, but also the amount that is properly recycled.
Overall, only 22.3% of global e-waste was properly recycled in 2022.
Although European countries produce the most e-waste per capita, they also recycle the largest proportion of this waste.
European countries recycled 7.53 kg of e-waste per person, while countries in the Americas recycled only 4.2 kg per person.
Meanwhile, in Africa, less than 1% of e-waste is officially listed as recycled.
Researchers estimate that 16 million tons of e-waste are managed in a large informal recycling system.
However, these informal systems also pose a huge risk to the health of workers and the local environment.
The report warns that electronic waste with a plug or battery should be considered an environmental hazard.
These contain toxic chemicals such as mercury that can cause irreversible brain damage in people exposed to them.


E-waste like these laptops discarded at a German recycling center contained metals worth an estimated £90 billion (£71 billion) in 2022.
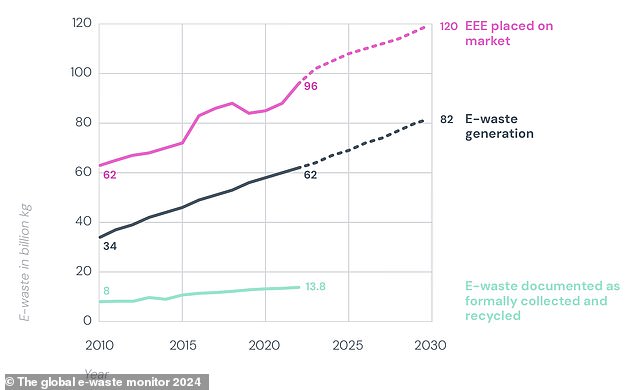

As this chart shows, e-waste generation (black) will continue to exceed recycling capacity (green) in the future.
Yet even with the help of informal recycling, researchers predict that the proportion of e-waste recycled each year will actually decrease in the future.
By 2023, researchers suggest only 20% of e-waste will be recycled, as waste growth continues to outpace global recycling efforts.
Cosmas Luckyson Zavazana, Director of the ITU Telecommunication Development Bureau, said: “The latest research shows that the global challenge posed by e-waste will only increase.
“With less than half of the world’s population implementing and enforcing approaches to manage the problem, this raises the alarm for strong regulations to increase collection and recycling.


E-waste contributes to a growing ecological problem as it pollutes the environment. Here, in a now-demolished scrapyard in Ghana, you can see the devastation of what were once thriving wetlands.
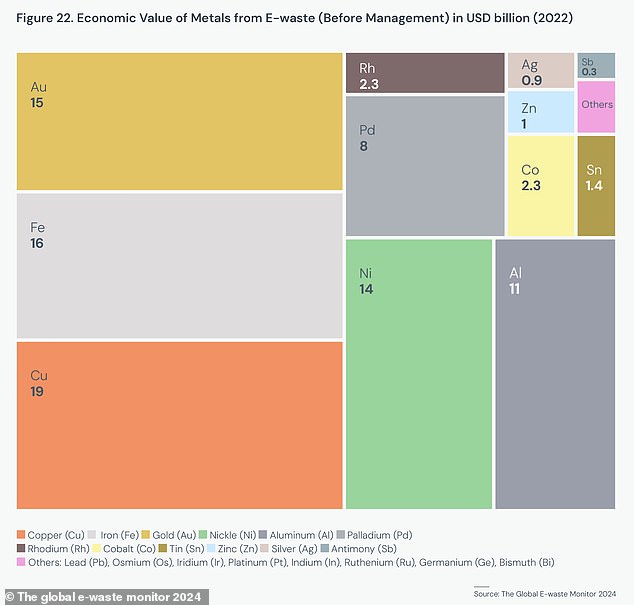

This chart shows the economic value of different minerals found in e-waste in 2022. Copper, iron and gold were the most valuable minerals that could have been saved through recycling.
As e-waste grows faster than our recycling capacity, this not only creates an environmental risk in itself, but also wastes billions of dollars worth of minerals.
Total e-waste produced in 2022 contained more than $90 billion (£71 billion) of metals, including $19 billion (£15 billion) of copper, $15 billion (£12 billion) of ) of gold and $16 billion (£12.6 billion) of iron. .
The UN report estimates that $62 billion (£49 billion) of recoverable natural resources were lost in 2022 alone.
If countries could increase their e-waste collection and recycling rates to just 60% by 2030, experts estimate the benefits would exceed costs by $38 billion (£30 billion).
However, the researchers write that these efforts are hampered by limited repair options, shorter product life cycles and poor management infrastructure.
