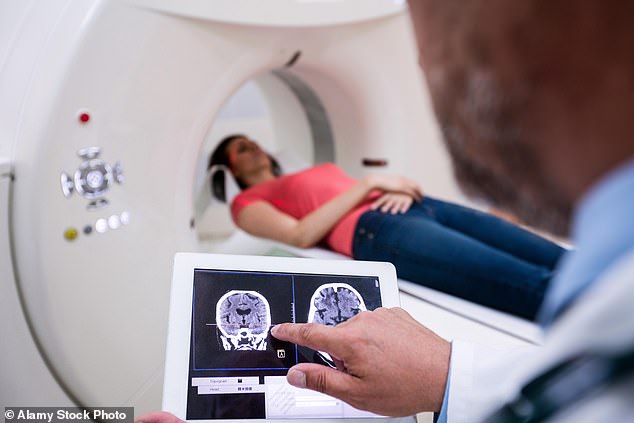
Researchers have developed a method for diagnosing autism that could save families years of uncertainty and spur crucial early treatment.
New AI analysis can identify genetic markers of autism through biological activity in the brain, they report, with 89 to 95 percent accuracy.
This new method begins with standard brain mapping via magnetic resonance imaging (MRI) before reanalyzing those scans via AI to detect the movements of proteins, nutrients and other processes within the brain that may indicate autism.
“Autism is traditionally diagnosed through behaviour” – through a person’s speech, for example – as noted by the medical team behind the process. “But it has a strong genetic basis.”
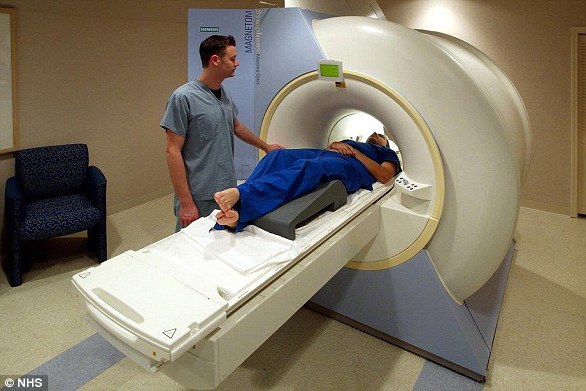
A new method for diagnosing autism starts with standard brain mapping via magnetic resonance imaging, or MRI (pictured above), but reanalyzes those scans via AI to detect the movements of proteins, nutrients and other processes in the brain that indicate the condition.
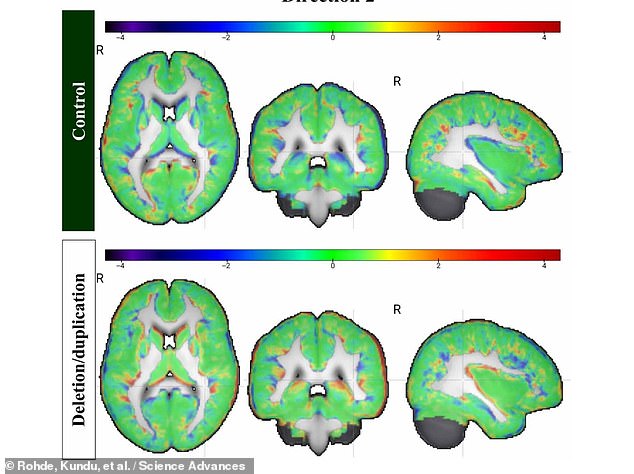
Above, brain scan data comparing a control brain without autism (top row) to a brain containing deletions or duplications of genetic material linked to autism (bottom row). The “noise” in this scan data was reduced on a three-dimensional pixel-by-pixel basis using the “Z-score mapping” method.
Currently, autism affects one in 36 children, according to the Centers for Disease Control and PreventionWhich means that more than 90,000 children are born with this developmental disorder each year in the U.S.
But autism is notoriously difficult to detect and the vast majority of children with the condition will not be diagnosed. They are diagnosed up to five years of age and present clear behavioral signs.
Worse, that identification process often involves years of uncertainty, dozens of trips to the hospital and a battery of tests — including speech and language screenings, observational interviews and more — that can be stressful for children and families.
Researchers hope the new diagnostic technique will soon enable doctors to pinpoint more specific genes responsible for autism, first revealing the actual biological pathways through which autism changes the way the brain grows and functions.
As a spokesperson for a university that supports the new method put it, the method ‘cracks the code of autism’, although it is not yet known when it might become common use.
Dr. Shinjini Kunduassistant professor of radiology at Washington University in St. Louis, developed this new technique of artificial intelligence and mathematical modeling of the brain with machine learning while she was a graduate student and researcher.
The method, called “transport-based morphometry” after the transport of biological matter in the brain, focuses on identifying patterns linked to key fragments of the genetic code.
These genetic code sequences, called “copy number variations” (CNVs), reveal segments of DNA that have been deleted or duplicated — alterations that have been linked to autism in previous research.
“Some copy number variations (CNVs) are known to be associated with autism,” according to biomedical engineering professor Dr. Gustavo Rohde, who taught Dr. Kundu during her PhD studies.
“But its link to brain morphology — in other words, how different types of brain tissue, such as gray or white matter, are organized in our brains — is not well known,” said Dr. Rohde, who now teaches at the University of Virginia.
“Discovering how CNV relates to brain tissue morphology,” he explained, “is an important first step toward understanding the biological basis of autism.”
Drs. Kundu and Rohde, and their collaborators from the Department of Neurology at the University of California, San Francisco, published their results of developing this new method of identifying autism in June in the journal Scientific advances.
Participants in the nonprofit Simons Individual Variation Project, a cohort of subjects with known genetic variations linked to autism, provided key data used in the new study.
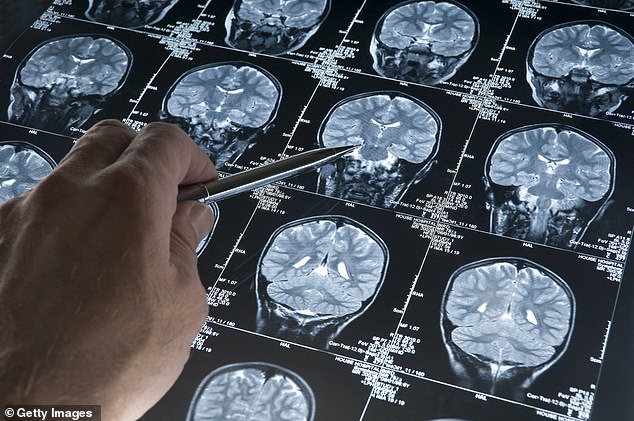
“Discovering how CNV (deletion or duplication of the genetic code) relates to brain tissue morphology,” said study co-author Dr. Gustavo Rohde, “is an important first step toward understanding the biological basis of autism.” The new method identifies these changes in brain morphology
The researchers recruited their “control subjects” from other medical or clinical settings based on their similarities to Simons’ group (such as same age, sex, and nonverbal IQ), to reduce variables that might confound their results.
According to Rohde, most machine learning methods that analyze medical data, such as MRI scans, do not incorporate a mathematical model for the numerous biological processes that are hidden in that data.
In contrast, previous AI models only looked for patterns to identify abnormalities or statistical anomalies in the health data of various patients.
However, Dr. Kundu’s “transport-based morphometry” could help researchers discern even more revealing biological variations within brain structures, beyond the deletions or duplications associated with CNVs and autism.
Given the reports that 90 percent of all medical data comes from similar images, the team hopes this method can help extract useful new information from old tools.
“Important discoveries may be possible from such large amounts of data if we use more appropriate mathematical models to extract such information,” said Dr. Rohde.
“We hope that the findings,” he added, “can point to brain regions and eventually mechanisms that can be harnessed for therapies.”