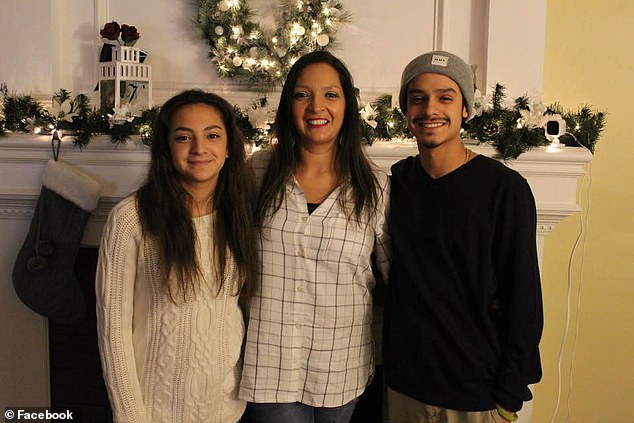The family of DJ Lisa López-Galván killed during Wednesday’s mass shooting in Kansas City said she died at the scene and not in a hospital as previously reported.
His statement was delivered Thursday by local Westside resident Beto Lopez, as Missouri police continue to investigate what happened at the devastated Super Bowl celebration.
As of this writing, two suspects are currently in custody, and Lopez claimed Thursday that his 44-year-old sister, and beloved local DJ, was gunned down by at least one of them.
López, a respected figure in the city’s Hispanic community, revealed that his nephew, López-Galván’s son and his 19-year-old niece were present and witnessed the ruthless murder of his mother.
When he was in his early 20s, López Galván’s son was also shot, López said, stating that it was his nephew who was rushed to a hospital and not his sister.
He added that two other relatives, two primary school girls, were also beaten, while police said nine children were hospitalized. The investigation into him remains ongoing as 13 other people were injured.
Scroll down to watch the video:
Lisa López-Galván, seen with her two children, Adrianna, 19, and Marc, 20, died at the scene of the shooting at the Kansas City Chiefs Super Bowl parade, police and family revealed. Her two children were also present, and Marc was shot in the leg, his relatives said.

Her young cousins, identified only as the elementary school-aged daughters of Kansas City resident Erika Reyes, were also present when the initial gunshots were heard and both were hit by gunfire, although Lopez said their wounds were “not life-threatening.” their lives”.
“She never made it to the hospital,” Lopez said of her younger sister’s fate, a statement corroborated by police on Thursday, who confirmed that she died at the scene, contrary to previous reports.
Other than that, the executive director of a local youth center said the rest is still unclear and it was unclear whether the children’s father, Mike Galvan, 66, was with the party.
From what he knows so far, he said that his sister was with her son Marc López-Galván and her daughter Adriana.
Her young cousins, identified only as relative Erika Reyes’ elementary school-aged daughters, were also there on the west side of Union Station when the initial shots rang out.
Sadly, everyone except Adriana was hit, he said, and her little sister was fatally wounded.
‘They were enjoying the day and getting ready to go home. “Everyone was,” Lopez lamented, offering the crucial information to the Kansas City Star.
They were all together. They were caught in the crossfire that was occurring.
He went on to reveal the extent of the young man’s injuries, stating that his twenty-something nephew was hit in the leg and has since been released from the hospital.
His sister, although not physically injured, is struggling mentally, he said, before expressing concern for both of them when they “witnessed their mother being shot.”

The claim was relayed Thursday by local Westside resident Beto Lopez, as police in Missouri continue to investigate what happened at the devastated Super Bowl celebration.

“She never made it to the hospital,” Lopez said of her younger sister’s fate, a statement backed up by police on Thursday, who confirmed that she died at the scene, contrary to previous reports.

López-Galván, who worked as a DJ at a community radio station, leaves behind his two children,

Lisa López-Galván, pictured here with her husband Mike Galván, died during surgery at a hospital from a gunshot wound to the abdomen. It is unclear if her spouse was also there during the attack.
Although his son has been released from the hospital, the executive director of Westside’s Guadalupe Center said the same applies to him, telling the local newspaper, “We’re going to have to really help him mentally to get through this.”
And he continued: “In addition to the shock, the initial shock, (Marc) was there, with his mother, when all this happened.”
Lopez further revealed how his sister, a disc jockey for a Hispanic music show on community radio station KKFI, was the youngest of four siblings, including himself.
“Of the four of us siblings, she was the life of the party,” he said. ‘Joyful person, I never met a stranger. She was well-liked in the community and she had a big heart.’
She added that the mother of two had also been a die-hard Kansas City fan and was thrilled when they won the big game.
No plans have been made for the funeral, he said, before revealing the current condition of the two girls hit by the hail of bullets.
“(Their injuries) are not life-threatening,” Lopez told The Star.
He added that the family has not yet seen his sister’s body, because it is part of an active homicide investigation.
“When you have a… It’s murder,” he said, before falling silent. ‘Even for me to say that, ‘My sister was murdered,’ (it’s) absurd to even say that.’

Images circulating on social media show a group of people detained after the shooting, some of whom appear to be minors. It is unclear if those in the photo are suspects.

A man dressed in a red tracksuit was quickly detained after the shooting, although it is unclear if he was involved in the tragedy.

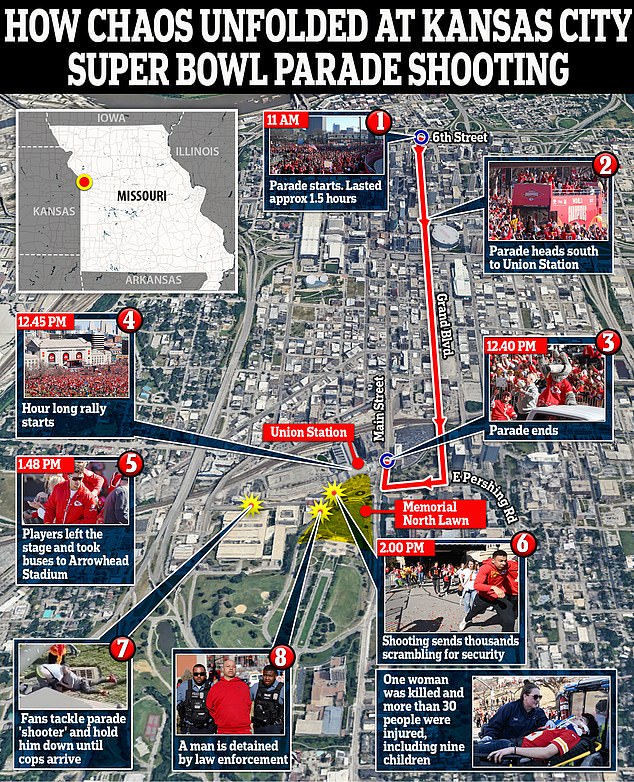
Terrified parade-goers fled for their lives after gunmen opened fire shortly after the Superbowl winners walked off the stage.
He went on to explain how ‘when there is an investigation that is still active, until the investigation is over they will give us some details on how we can recover his body for a respectful burial,’ before explaining how they plan to honor his memory.
“We’re going to try to make sure she’s not forgotten,” he said, as police confirmed they had detained two juvenile suspects.
“If we have to use this as an example to finally get proper attention in Congress, to really address gun violence,” he continued. ‘That’s something that will be part of my mission for the rest of my life.
“She was (just) trying to go, like everyone else, to celebrate.”
The important count came shortly after Children’s Mercy hospital staff also confirmed they had treated nine children for gunshot wounds in the incident.
Everyone, including Reyes’ young daughters, is expected to recover, they said, and police officers added that of the 22 injured victims, some of whom were trampled in the melee, half were children under 16 years old.
Police Chief Stacey Graves added that while her department still needs to review the evidence, the victims were between eight and 47 years old. Lopez did not specify the age of the two girls on Sunday, other than that they were in elementary school.
University Health officials added that of the adult victims with gunshot wounds who were rushed to their hospital, two remain in critical condition and in the ICU.
However, medical staff said both are showing signs of improvement, while Graves revealed that of the three people detained after Wednesday’s incident, two are still detained.
Both are teenagers and a third youth was released sometime before his testimony Thursday.
He added that the 24-hour investigative hold rule does not apply to minors, meaning officials can hold them much longer before losing them.
His investigation, like Friday, remains ongoing.